Example
In this simple demonstration, you will see how to calculate ZBLMIp (Z score of the corrected MIp using BLOSUM62 pseudo frequencies) for a Pfam MSA from the Julia REPL or using a MIToS script in the system command line.
MSA from the Julia REPL or using a MIToS script in the system command line.
MIToS in the Julia REPL
If you load the Pfam module from MIToS, you will get access to a set of functions that work with Pfam MSAs. In this case, we are going to use it for download a Stockholm MSA from the Pfam website and read it into Julia.
MSA from the Pfam website and read it into Julia.
using MIToS.Pfam
pfam_file = downloadpfam("PF10660")
msa = read_file(pfam_file, Stockholm, generatemapping = true, useidcoordinates = true)AnnotatedMultipleSequenceAlignment with 3070 annotations : 1521×64 Named Matrix{MIToS.MSA.Residue}
Seq ╲ Col │ 31 34 35 36 37 … 131 132 133 134 135
─────────────────────────┼────────────────────────────────────────────────────
A0A6J3Q5A2_TURTR/9-44 │ - - - - - … K D H R -
A0A8I3PMH8_CANLF/15-57 │ - - - - - N Q K Q Q
A0A7K7LDH8_9AVES/2-65 │ - E S L A K K K Q Q
A0A7K9L2R5_9PASS/2-65 │ - E T L A K K K Q Q
A0A851JM88_9PASS/4-38 │ - - - - - K D K C -
A0A7L0KET4_CHATO/4-38 │ - - - - - K D K C -
A0A091W6B3_OPIHO/2-35 │ - - - - - K D K C -
A0A2K6GIS2_PROCO/9-41 │ - - - - - K D H R -
⋮ ⋮ ⋮ ⋮ ⋮ ⋮ ⋱ ⋮ ⋮ ⋮ ⋮ ⋮
A0A7K9YJI6_9GALL/2-65 │ - E S L A K K K Q Q
CID2A_XENLA/1-66 │ M E S I A K K K Q Q
A0A9W2WD00_PHYMC/10-44 │ - - - - - K D H R -
A0A852JHR7_SPIPA/3-36 │ - - - - - K D K - -
A0A6I9HMT6_GEOFO/9-41 │ - - - - - K D K - -
A0A183P1E2_9TREM/1-64 │ M N I L H H L G K -
A0A669CBP1_ORENI/1-66 │ - E T I A K K K K Q
A0A8C4MK81_EQUAS/5-39 │ - - - - - … K D H C NThe keyword argument generatemapping of read_file allows to generate sequence and column mappings for the MSA. Column mapping is the map between of each column on the MSA object and the column number in the file. Sequence mappings will use the start and end coordinates in the sequence ids for enumerate each residue in the sequence if useidcoordinates is true.
You can plot this MSA and other MIToS’ objects using the Plots package. The installation of Plots is described in the Installation section of this site:
package. The installation of Plots is described in the Installation section of this site:
using Plots
plot(msa)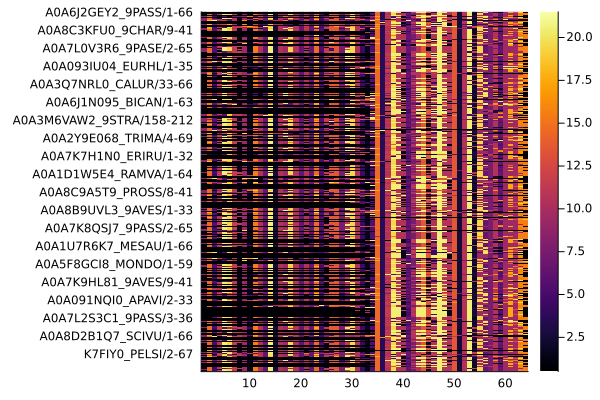
The Information module of MIToS has functions to calculate measures from the Information Theory , such as Shannon Entropy and Mutual Information (MI), on a MSA. In this example, we will estimate covariation between columns of the MSA with a corrected MI that use the BLOSUM62 matrix for calculate pseudo frequencies (
, such as Shannon Entropy and Mutual Information (MI), on a MSA. In this example, we will estimate covariation between columns of the MSA with a corrected MI that use the BLOSUM62 matrix for calculate pseudo frequencies (BLMI).
using MIToS.Information
ZBLMIp, BLMIp = BLMI(msa)
ZBLMIp # shows ZBLMIp scores36×36 Named PairwiseListMatrices.PairwiseListMatrix{Float64, false, Vector{Float64}}
Col1 ╲ Col2 │ 67 68 … 133 134
────────────┼──────────────────────────────────────────────────────────
67 │ NaN 0.0927656 … 0.312175 -0.0747719
68 │ 0.0927656 NaN -0.00813555 -0.188341
69 │ -0.0975533 -0.0237266 -0.0226986 -0.103186
71 │ 0.00863921 -0.00597914 0.0115664 0.101111
73 │ 0.0521814 0.00416266 -0.0131909 0.240317
87 │ 0.0400599 0.0428923 0.0979899 -0.0359407
93 │ 0.0318728 -0.025663 -0.0756465 0.0315444
101 │ 0.0153942 -0.0402377 0.0895165 0.120163
⋮ ⋮ ⋮ ⋱ ⋮ ⋮
127 │ 0.011762 0.0915685 -0.116568 0.0813957
128 │ -0.0176941 0.0170251 0.239387 0.0232993
129 │ 0.145628 0.166716 -0.0600544 -0.229281
130 │ -0.0954998 -0.0610818 -0.161604 -0.0259002
131 │ 0.0891384 0.0324601 0.126517 -0.0500363
132 │ 0.0731305 -0.104509 -0.0734673 -0.107737
133 │ 0.312175 -0.00813555 NaN -0.0685581
134 │ -0.0747719 -0.188341 … -0.0685581 NaNOnce the Plots package is installed and loaded, you can use its capabilities to visualize this results:
heatmap(ZBLMIp, yflip = true, c = :grays)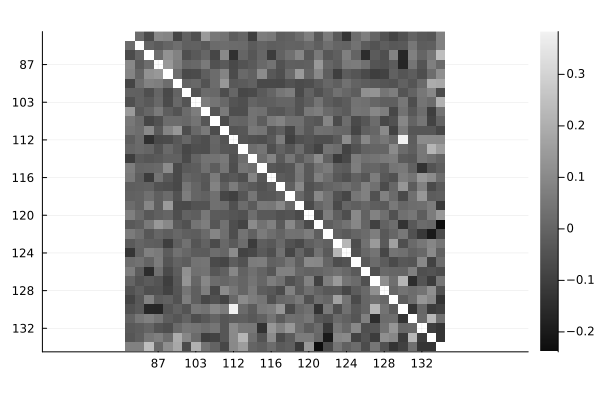
MIToS in system command line
Calculate ZBLMIp on the system shell is easy using the script called BLMI.jl in the MIToS_Scripts.jl package. This script reads a MSA file, and writes a file with the same base name of the input but with the
package. This script reads a MSA file, and writes a file with the same base name of the input but with the .BLMI.csv extension.
julia BLMI.jl PF14972.stockholm.gz